Issue 67: A new resource for legume diversity and systematics: The Nit-Fix Project
Heather Kates1, Ryan Folk2, Robert Guralnick1, Doug Soltis1, Pamela Soltis1
1 Florida Museum of Natural History, University of Florida
2 Department of Biological Sciences, Mississippi State University
Legumes are famous for their symbiotic relationship with bacteria that fix nitrogen, but we still have much to learn about this globally important symbiosis. Understanding the incredible diversity of legume species and their relatives is one key to unlocking the mystery of the evolutionary origins and genetic basis of this critical innovation in plant and microbial evolution.
Nitrogen-fixing symbioses between plants like legumes and their relatives can give plants an advantage in nitrogen-poor soils, and may be associated with ecological shifts to extreme habitats. Deconstructing how this symbiosis works across a wide range of plant diversity could reduce fertilizer use, thereby reducing the high energy cost of production, limiting agricultural runoff, and securing the food supply in arid parts of the globe, while enriching our understanding of how symbioses have been critical planetary drivers of evolution and ecology.
We, along with colleagues at the University of Florida and the University of Wisconsin-Madison, have recently been awarded grants from the U.S. Department of Energy and the U.S. National Science Foundation to investigate the evolutionary origins of nitrogen-fixing symbioses and identify the genomic innovations that led to root nodule development. The results of these analyses will be applied to verifying the molecular mechanisms of nodulation and engineering nodulation in bioenergy crops (https://nitfix.org).
Any exploration of a trait’s genetic basis and evolution relies on a robust phylogenetic hypothesis. Because of the explosive diversification of plants that acquired the nodular nitrogen-fixing symbiosis, all of which are members of the “the nitrogen-fixing clade” of angiosperms, building the phylogenetic framework needed to rigorously answer questions about this symbiosis requires unprecedented efforts to reconstruct relationships among legumes and other members of the nitrogen-fixing clade (non-legume Fabales, Rosales, Cucurbitales, and Fagales).
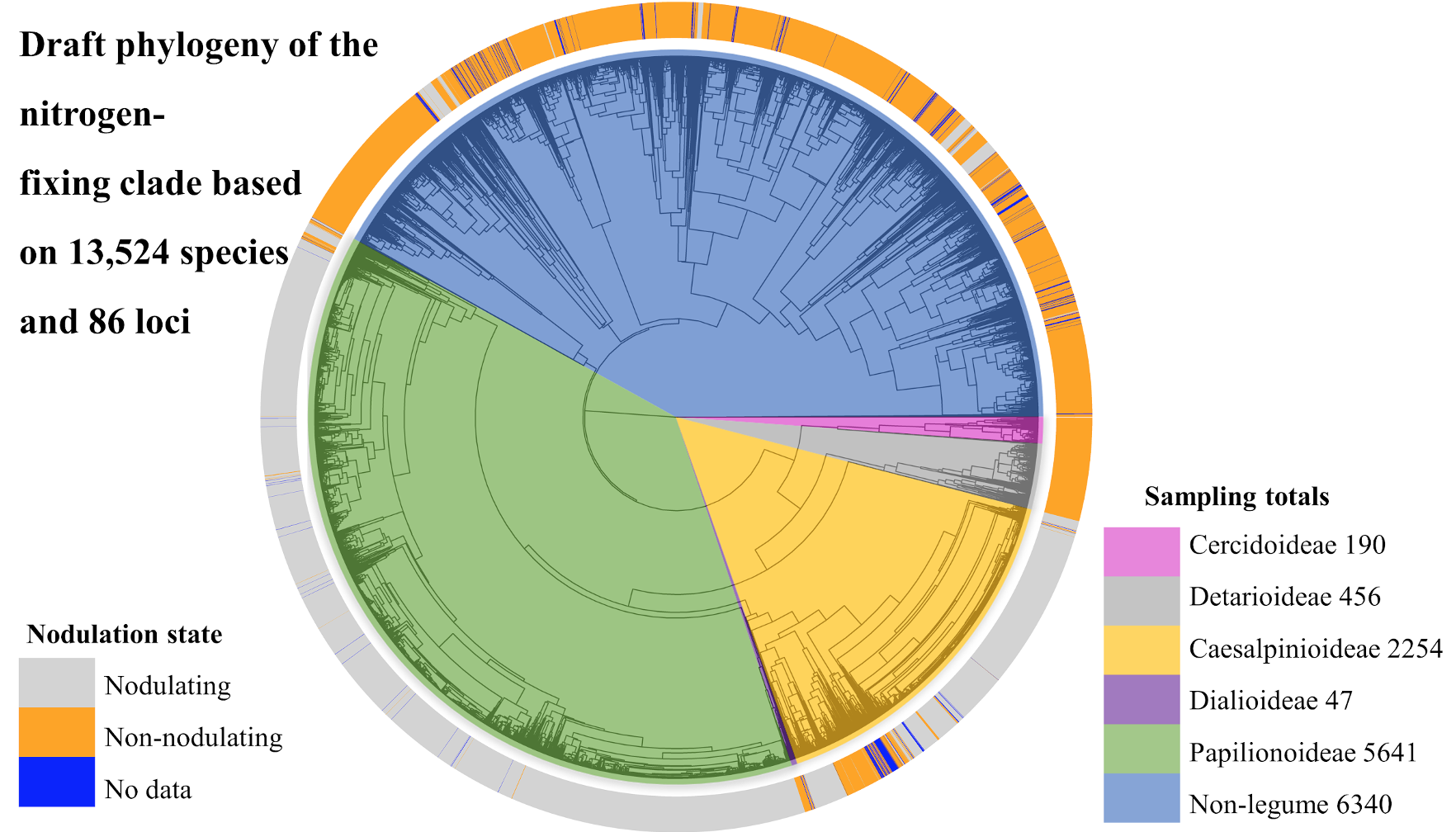
We have assembled phylogenomic data comprising hundreds of gene sequences for over 13,000 species in the nitrogen-fixing clade as part of the wider “NitFix” project. We are using a subset of these gene sequences for phylogenetic analysis (Fig. 1). The rest of the gene set comprises functional genes related to nodulation that we are screening for association with nodulation.
Using plant specimens preserved in herbaria, we have sampled and sequenced close to 50% of the diversity of the nitrogen-fixing clade, including over 8,000 legume species, some of which have never been included in a molecular study. In collaboration with experts of the Legume Phylogeny Working Group, our evolutionary analyses (Fig. 1) are based on the most up-to-date legume taxonomy and genus-level nodulation-trait information available, both of which represent critical improvements upon earlier efforts to study the evolution of nodulation in the nitrogen-fixing clade.
Figure 1. A draft phylogeny of the nitrogen-fixing clade based on currently assembled data and analysis. Relationships among clades may change based on on-going analyses. Genus-level nodulation states are plotted at the tree tips, and legume subfamily clades and numbers of species sampled per clade indicated with colors (the monospecific subfamily Duparquetioideae sample is not indicated).
Our dataset will form an integral resource for the study of the evolution of nodulation and will make the nodulating clade the most densely sampled model clade for comparative research in plants. We also hope that our dataset will be a valuable resource for the legume research community to investigate compelling questions related to legume systematics and evolution due to its increased sampling and resolution of many legume genera and its potential to robustly resolve deeper relationships within legume phylogeny.
For more information on this project, please visit https://nitfix.org or email hkates@ufl.edu or rfolk@biology.msstate.edu