Coordinators: Joe Miller (Global Biodiversity Information Facility) Copenhagen Denmark and Vanessa Terra Universidade Federal de Uberlândia - UFU
Introduction
Open Tree of Life aims to construct a comprehensive, dynamic and digitally-available tree of life by synthesizing published phylogenetic trees along with taxonomic data. We think of it as the phylogenetic version of GBIF. GBIF combines data from thousands of herbarium datasets into a system that allows anyone to access occurrence data in a geographic area or of a taxonomic group. Likewise OToL, since we don’t have a single analyzed phylogenetic tree of all life, does the same with open access data. OToL users can download a subset tree, for example for all of Leguminosae (Fabaceae) or from a list of species. As with GBIF, the better the input data, the better the output data. OToL trees can be used for many purposes, including research and for visualizations.
It is important to recognize that the OToL synthetic tree includes terminals from published phylogenetic analyses from many sources, and thus include various genes, morphology, and sampling. In addition, many terminals in the tree are known only from the taxonomy, that is they are not currently represented in a source phylogeny currently in OToL. OToL uses a robust Taxonomic Name Resolution Service (TNRS) to place these terminals. For example, the Fabaceae clade (OTT: 560323) includes 24,479 species but only 4,835 species are placed based on input phylogenies. Species that are not in any input trees are placed as a polytomy at the genus level. To resolve these polytomies, we need to input as many high quality trees as possible. It is also possible through OToL to download a tree that only includes terminals that are from phylogenetic analyses. For information about OToL see the about tab here.
Emily Jane McTavish (UC Merced and an OToL PI) has sent us information, including the Leguminosae synthetic trees and lists below. Our objective is to work with Emily Jane McTavish to improve the Leguminosae clade of OToL. Because it is very easy to import a phylogeny from Treebase into OToL for further curation, harvesting legume Treebase studies is the first place to improve the OToL synthetic tree.
Status of Legumes in OToL
| ToL Derived Phylogeny (version Dec 2019) | # of Terminals | link to file |
|---|---|---|
| All Leguminosae species | 24,479 | at OToL |
| Only based on phylogenies | 4,835 | Newick |
-
Published studies currently integrated into current OTol versions
-
Partially curated studies in OToL that contain Leguminosae species
This google spreadsheet lists the status of Leguminosae trees in OToL (January 2021) and indicates target trees to import from published studies:
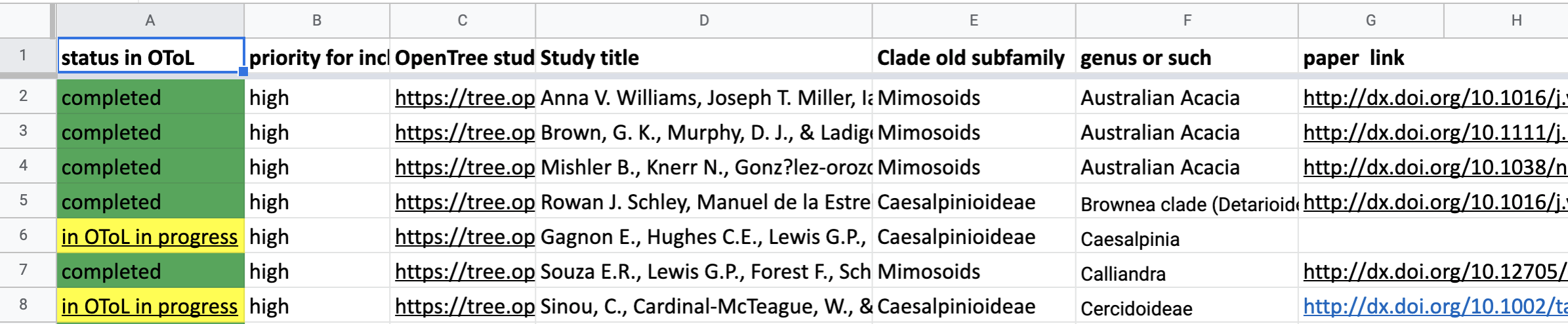
| Tab | Status | Action |
|---|---|---|
| OToL List | Completed | Already in OToL, no action needed |
| OToL List | In Progress | Considering your knowledge of the clade and paper, is this important to include? |
| OToL List | Priority | Update Priority in column 2 |
| Potential Trees | In Treebase | Import into OToL from TreeBase and curate |
| Potential Trees | pdf, not in treebase | Determine if priority, find tree or contact authors, or determineif newer studies cover this clade |
| Potential Trees | to assess | Is this worth chasing? or are there newer studies that cover this clade |
| Potential Trees | find pdf | Is this worth chasing? or are there newer studies that cover this clade |
| Suggest a tree | Trees needed | Recent trees that cover groups not in current synthetic tree, especially species level |
/* Potential Trees: This list of trees was obtained mailnly from LPWG citations. This is an old list and needs updating.
Priority list of things to do by the Working Group
- Identify studies that are high priority to include
- species level
- missing in current synthetic tree
- recent publications
- trees that resolve the backbone, new phylogenomic studies
- in Treebase or we can easily get Treefiles
- Identify trees from the OToL List and Potential Tree list that are worthy of inclusion
- Download the current OToL tree file Newick and find gaps in coverage
- Update the taxonomy in the spreadsheet
- to help find gaps
- to eliminate mistakes
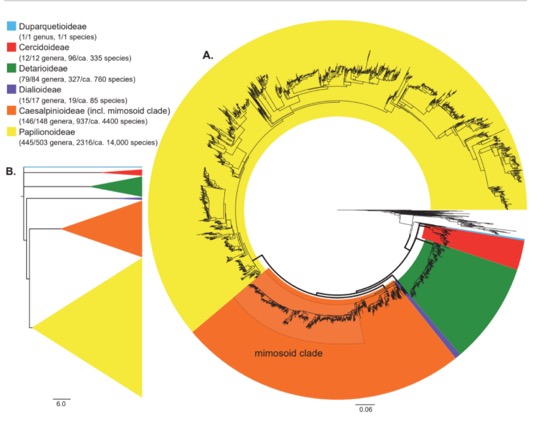
- Perhaps focus on Caesalpinioideae (incl. mimosoid clade), Dialioideae, Detarioideae, Cercidoideae and Duparquetioideae
- obtain a much better current coverage in these clades
Please help us!
How to curate files in OToL
Click on an OpenTree study link to see what the interface looks like. You will need a GitHub account to edit. More details to be added later. The OToL synthetic tree is only updated occasionally. The most recent was in May 2024.
Publication idea
We could rewrite a paper describing the before and after this project. How many species and studies did we add? How much better/useful is the OToL phylogeny? We could provide examples of how the tree would be used, especially in connection to the Taxonomic and Occurrences Working Groups. GBIF is working on a phylogenetic viewer that has been added to the Legume Data Portal.
Join the Working Group
Please send an email to Joe Miller or Vanessa Terra or just start looking for studies to include. No experience in curating OToL files is necessary. The most important thing we need is your knowledge of the best trees to include.